Note
Go to the end to download the full example code.
ECoG Movement decoding example#
This example notebook read openly accessible data from the publication Electrocorticography is superior to subthalamic local field potentials for movement decoding in Parkinson’s disease (Merk et al. 2022 <https://elifesciences.org/articles/75126>_). The dataset is available here.
For simplicity one example subject is automatically shipped within this repo at the py_neuromodulation/data folder, stored in iEEG BIDS format.
from sklearn import metrics, model_selection, linear_model
import matplotlib.pyplot as plt
import py_neuromodulation as nm
Let’s read the example using mne_bids. The resulting raw object is of type mne.RawArray. We can use the properties such as sampling frequency, channel names, channel types all from the mne array and create the nm_channels DataFrame:
(
RUN_NAME,
PATH_RUN,
PATH_BIDS,
PATH_OUT,
datatype,
) = nm.io.get_paths_example_data()
(
raw,
data,
sfreq,
line_noise,
coord_list,
coord_names,
) = nm.io.read_BIDS_data(PATH_RUN=PATH_RUN)
channels = nm.utils.set_channels(
ch_names=raw.ch_names,
ch_types=raw.get_channel_types(),
reference="default",
bads=raw.info["bads"],
new_names="default",
used_types=("ecog", "dbs", "seeg"),
target_keywords=["MOV_RIGHT"],
)
Extracting parameters from /opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/data/sub-testsub/ses-EphysMedOff/ieeg/sub-testsub_ses-EphysMedOff_task-gripforce_run-0_ieeg.vhdr...
Setting channel info structure...
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/utils/io.py:61: RuntimeWarning: Did not find any events.tsv associated with sub-testsub_ses-EphysMedOff_task-gripforce_run-0.
The search_str was "/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/data/sub-testsub/**/ieeg/sub-testsub_ses-EphysMedOff*events.tsv"
raw_arr = read_raw_bids(bids_path)
Reading channel info from /opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/data/sub-testsub/ses-EphysMedOff/ieeg/sub-testsub_ses-EphysMedOff_task-gripforce_run-0_channels.tsv.
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/utils/io.py:61: RuntimeWarning: Other is not an MNE-Python coordinate frame for IEEG data and so will be set to 'unknown'
raw_arr = read_raw_bids(bids_path)
Reading electrode coords from /opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/data/sub-testsub/ses-EphysMedOff/ieeg/sub-testsub_ses-EphysMedOff_space-mni_electrodes.tsv.
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/utils/io.py:61: RuntimeWarning: There are channels without locations (n/a) that are not marked as bad: ['MOV_RIGHT']
raw_arr = read_raw_bids(bids_path)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/utils/io.py:61: RuntimeWarning: Not setting position of 1 misc channel found in montage:
['MOV_RIGHT']
Consider setting the channel types to be of EEG/sEEG/ECoG/DBS/fNIRS using inst.set_channel_types before calling inst.set_montage, or omit these channels when creating your montage.
raw_arr = read_raw_bids(bids_path)
This example contains the grip force movement traces, we’ll use the MOV_RIGHT channel as a decoding target channel. Let’s check some of the raw feature and time series traces:
plt.figure(figsize=(12, 4), dpi=300)
plt.subplot(121)
plt.plot(raw.times, data[-1, :])
plt.xlabel("Time [s]")
plt.ylabel("a.u.")
plt.title("Movement label")
plt.xlim(0, 20)
plt.subplot(122)
for idx, ch_name in enumerate(channels.query("used == 1").name):
plt.plot(raw.times, data[idx, :] + idx * 300, label=ch_name)
plt.legend(bbox_to_anchor=(1, 0.5), loc="center left")
plt.title("ECoG + STN-LFP time series")
plt.xlabel("Time [s]")
plt.ylabel("Voltage a.u.")
plt.xlim(0, 20)
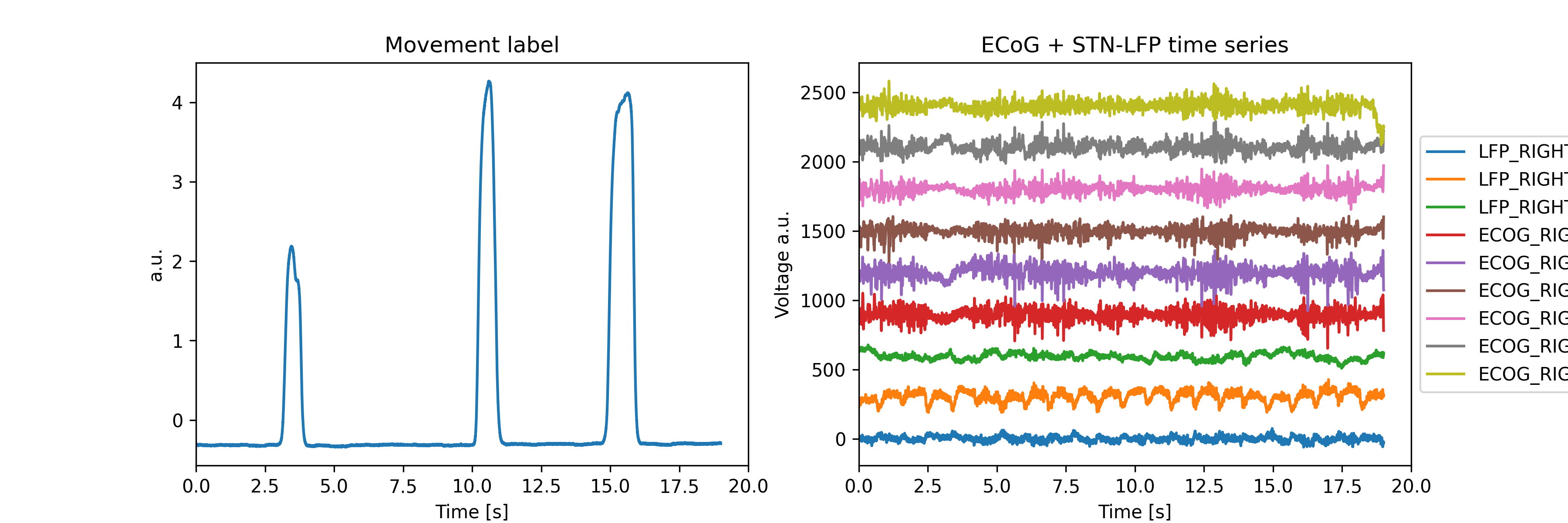
(0.0, 20.0)
settings = nm.NMSettings.get_fast_compute()
settings.features.welch = True
settings.features.fft = True
settings.features.bursts = True
settings.features.sharpwave_analysis = True
settings.features.coherence = True
settings.coherence_settings.channels = [["LFP_RIGHT_0", "ECOG_RIGHT_0"]]
settings.coherence_settings.frequency_bands = ["high beta", ]
settings.sharpwave_analysis_settings.estimator["mean"] = []
settings.sharpwave_analysis_settings.sharpwave_features.enable_all()
for sw_feature in settings.sharpwave_analysis_settings.sharpwave_features.list_all():
settings.sharpwave_analysis_settings.estimator["mean"].append(sw_feature)
stream = nm.Stream(
sfreq=sfreq,
channels=channels,
settings=settings,
line_noise=line_noise,
coord_list=coord_list,
coord_names=coord_names,
verbose=True,
)
Feature Analysis Movement#
The obtained performances can now be read and visualized using the analysis.Feature_Reader.
# initialize analyzer
feature_reader = nm.analysis.FeatureReader(
feature_dir=PATH_OUT,
feature_file=RUN_NAME,
)
feature_reader.label_name = "MOV_RIGHT"
feature_reader.label = feature_reader.feature_arr["MOV_RIGHT"]
feature_reader.feature_arr.iloc[100:108, -6:]
(181, 458)
feature_reader._get_target_ch()
'MOV_RIGHT'
feature_reader.plot_target_averaged_channel(
ch="ECOG_RIGHT_0",
list_feature_keywords=None,
epoch_len=4,
threshold=0.5,
ytick_labelsize=7,
figsize_x=12,
figsize_y=12,
)
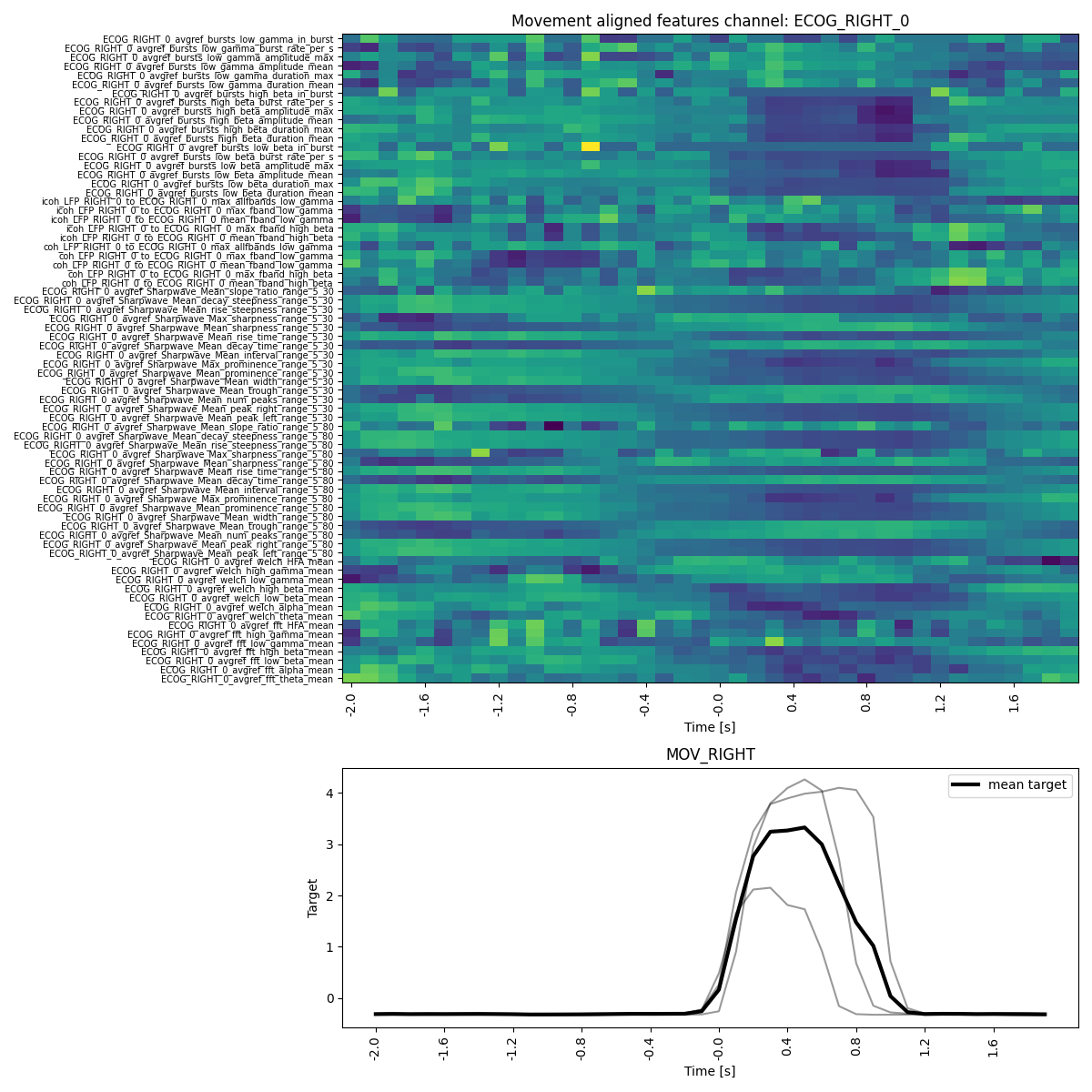
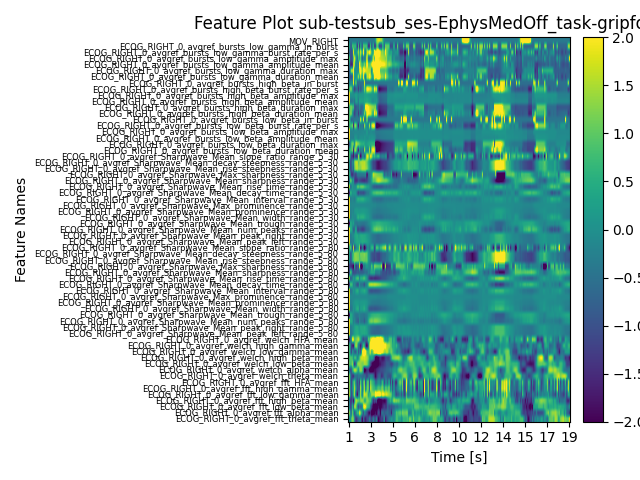
feature_reader.plot_all_features(
ytick_labelsize=6,
clim_low=-2,
clim_high=2,
ch_used="ECOG_RIGHT_0",
time_limit_low_s=0,
time_limit_high_s=20,
normalize=True,
save=True,
)
nm.analysis.plot_corr_matrix(
feature=feature_reader.feature_arr.filter(regex="ECOG_RIGHT_0"),
ch_name="ECOG_RIGHT_0_avgref",
feature_names=list(
feature_reader.feature_arr.filter(regex="ECOG_RIGHT_0_avgref").columns
),
feature_file=feature_reader.feature_file,
show_plot=True,
figsize=(15, 15),
)
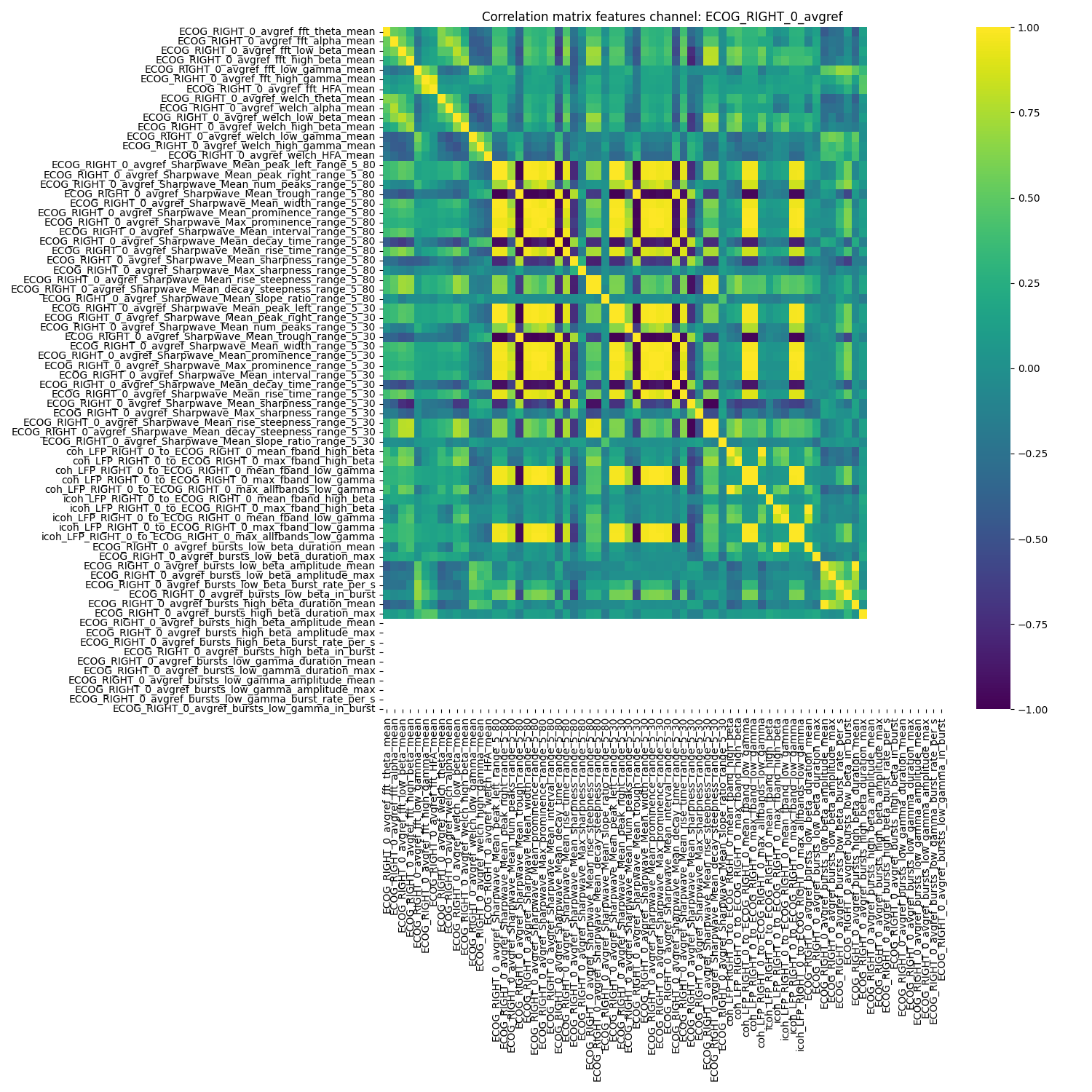
<Axes: title={'center': 'Correlation matrix features channel: ECOG_RIGHT_0_avgref'}>
Decoding#
The main focus of the py_neuromodulation pipeline is feature estimation. Nevertheless, the user can also use the pipeline for machine learning decoding. It can be used for regression and classification problems and also dimensionality reduction such as PCA and CCA.
Here, we show an example using a sklearn linear regression model. The used labels came from a continuous grip force movement target, named “MOV_RIGHT”.
First we initialize the Decoder class, which the specified validation method, here being a simple 3-fold cross validation,
the evaluation metric, used machine learning model, and the channels we want to evaluate performances for.
There are many more implemented methods, but we will here limit it to the ones presented.
model = linear_model.LinearRegression()
feature_reader.decoder = nm.analysis.Decoder(
features=feature_reader.feature_arr,
label=feature_reader.label,
label_name=feature_reader.label_name,
used_chs=feature_reader.used_chs,
model=model,
eval_method=metrics.r2_score,
cv_method=model_selection.KFold(n_splits=3, shuffle=True),
)
performances = feature_reader.run_ML_model(
estimate_channels=True,
estimate_gridpoints=False,
estimate_all_channels_combined=True,
save_results=True,
)
The performances are a dictionary that can be transformed into a DataFrame:
df_per = feature_reader.get_dataframe_performances(performances)
df_per
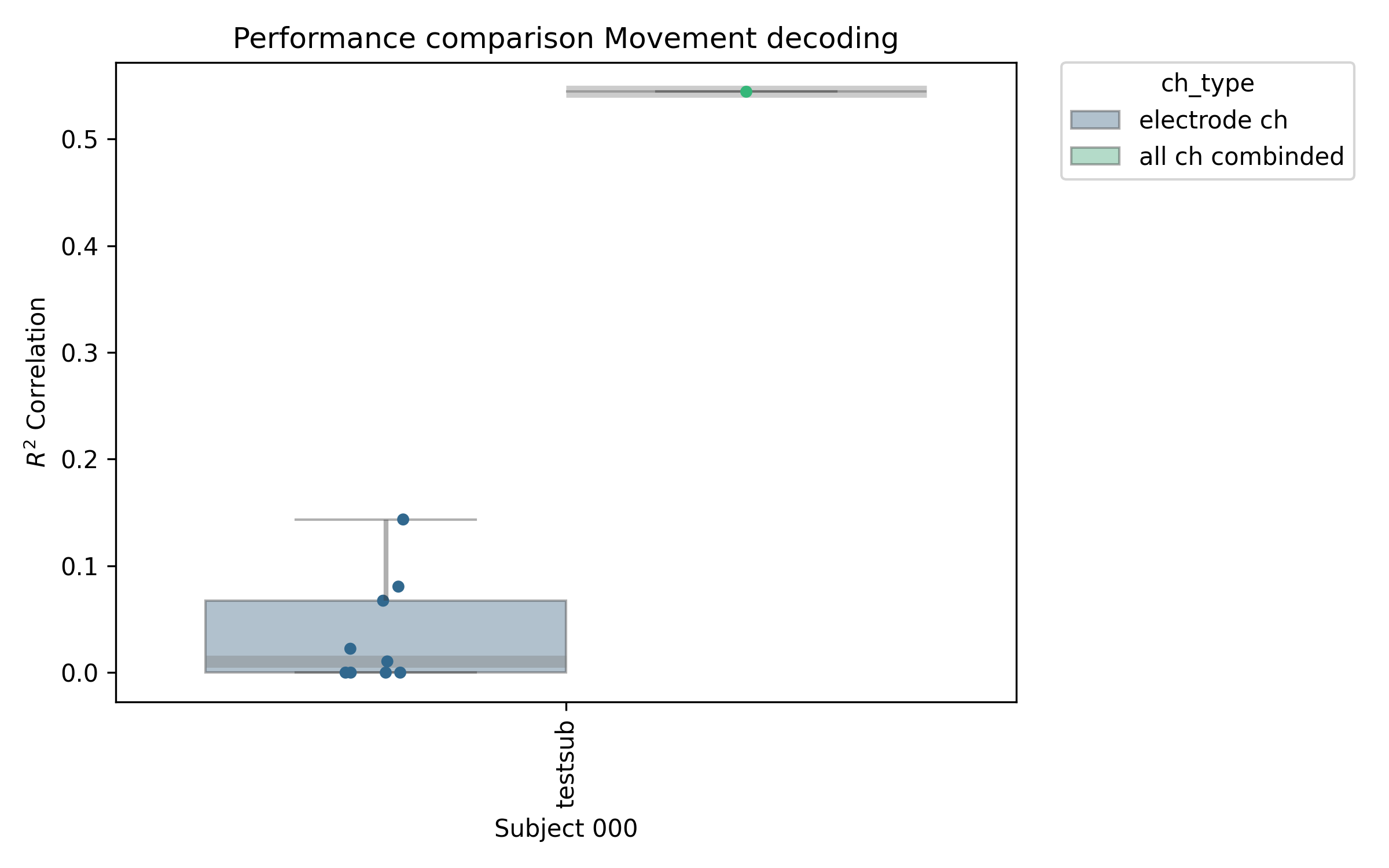
ax = nm.analysis.plot_df_subjects(
df_per,
x_col="sub",
y_col="performance_test",
hue="ch_type",
PATH_SAVE=PATH_OUT / RUN_NAME / (RUN_NAME + "_decoding_performance.png"),
figsize_tuple=(8, 5),
)
ax.set_ylabel(r"$R^2$ Correlation")
ax.set_xlabel("Subject 000")
ax.set_title("Performance comparison Movement decoding")
plt.tight_layout()
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/seaborn/categorical.py:700: PendingDeprecationWarning: vert: bool will be deprecated in a future version. Use orientation: {'vertical', 'horizontal'} instead.
artists = ax.bxp(**boxplot_kws)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/seaborn/categorical.py:700: PendingDeprecationWarning: vert: bool will be deprecated in a future version. Use orientation: {'vertical', 'horizontal'} instead.
artists = ax.bxp(**boxplot_kws)
Total running time of the script: (0 minutes 8.894 seconds)