Note
Go to the end to download the full example code.
First Demo#
This Demo will showcase the feature estimation and exemplar analysis using simulated data.
import numpy as np
from matplotlib import pyplot as plt
import py_neuromodulation as nm
Data Simulation#
We will now generate some exemplar data with 10 second duration for 6 channels with a sample rate of 1 kHz.
def generate_random_walk(NUM_CHANNELS, TIME_DATA_SAMPLES):
# from https://towardsdatascience.com/random-walks-with-python-8420981bc4bc
dims = NUM_CHANNELS
step_n = TIME_DATA_SAMPLES - 1
step_set = [-1, 0, 1]
origin = (np.random.random([1, dims]) - 0.5) * 1 # Simulate steps in 1D
step_shape = (step_n, dims)
steps = np.random.choice(a=step_set, size=step_shape)
path = np.concatenate([origin, steps]).cumsum(0)
return path.T
NUM_CHANNELS = 6
sfreq = 1000
TIME_DATA_SAMPLES = 10 * sfreq
data = generate_random_walk(NUM_CHANNELS, TIME_DATA_SAMPLES)
time = np.arange(0, TIME_DATA_SAMPLES / sfreq, 1 / sfreq)
plt.figure(figsize=(8, 4), dpi=100)
for ch_idx in range(data.shape[0]):
plt.plot(time, data[ch_idx, :])
plt.xlabel("Time [s]")
plt.ylabel("Amplitude")
plt.title("Example random walk data")
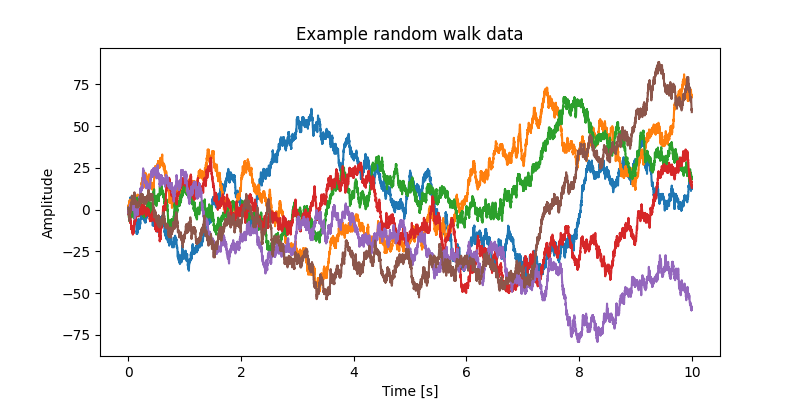
Text(0.5, 1.0, 'Example random walk data')
Now let’s define the necessary setup files we will be using for data preprocessing and feature estimation. Py_neuromodualtion is based on two parametrization files: the channels.tsv and the default_settings.json.
nm_channels#
The nm_channel dataframe. This dataframe contains the columns
Column name |
Description |
|---|---|
name |
name of the channel |
rereference |
different channel name for bipolar re-referencing, or average for common average re-referencing |
used |
0 or 1, channel selection |
target |
0 or 1, for some decoding applications we can define target channels, e.g. EMG channels |
type |
channel type according to the mne-python toolbox e.g. ecog, eeg, ecg, emg, dbs, seeg etc. |
status |
good or bad, used for channel quality indication |
new_name |
this keyword can be specified to indicate for example the used rereferncing scheme |
The nm_stream_abc can either be created as a .tsv text file, or as a pandas
DataFrame. There are some helper functions that let you create the
nm_channels without much effort:
nm_channels = nm.utils.get_default_channels_from_data(data, car_rereferencing=True)
nm_channels
Using this function default channel names and a common average re-referencing scheme is specified. Alternatively the define_nmchannels.set_channels function can be used to pass each column values.
nm_settings#
Next, we will initialize the nm_settings dictionary and use the default settings, reset them, and enable a subset of features:
settings = nm.NMSettings.get_fast_compute()
The setting itself is a .json file which contains the parametrization for preprocessing, feature estimation, postprocessing and definition with which sampling rate features are being calculated. In this example sampling_rate_features_hz is specified to be 10 Hz, so every 100ms a new set of features is calculated.
For many features the segment_length_features_ms specifies the time dimension of the raw signal being used for feature calculation. Here it is specified to be 1000 ms.
We will now enable the features:
fft
bursts
sharpwave
and stay with the default preprcessing methods:
notch_filter
re_referencing
and use z-score postprocessing normalization.
settings.features.fooof = True
settings.features.fft = True
settings.features.bursts = True
settings.features.sharpwave_analysis = True
We are now ready to go to instantiate the Stream and call the run method for feature estimation:
stream = nm.Stream(
settings=settings,
channels=nm_channels,
verbose=True,
sfreq=sfreq,
line_noise=50,
)
features = stream.run(data, save_csv=True)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/py_neuromodulation/features/fooof.py:74: DeprecationWarning:
The `fooof` package is being deprecated and replaced by the `specparam` (spectral parameterization) package.
This version of `fooof` (1.1) is fully functional, but will not be further updated.
New projects are recommended to update to using `specparam` (see Changelog for details).
from fooof import FOOOFGroup
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/fooof/core/funcs.py:67: RuntimeWarning: invalid value encountered in log10
ys = ys + offset - np.log10(knee + xs**exp)
Feature Analysis#
There is a lot of output, which we could omit by verbose being False, but let’s have a look what was being computed.
We will therefore use the nm_analysis class to showcase some functions. For multi-run -or subject analysis we will pass here the feature_file “sub” as default directory:
analyzer = nm.FeatureReader(
feature_dir=stream.out_dir, feature_file=stream.experiment_name
)
Let’s have a look at the resulting “feature_arr” DataFrame:
analyzer.feature_arr.iloc[:10, :7]
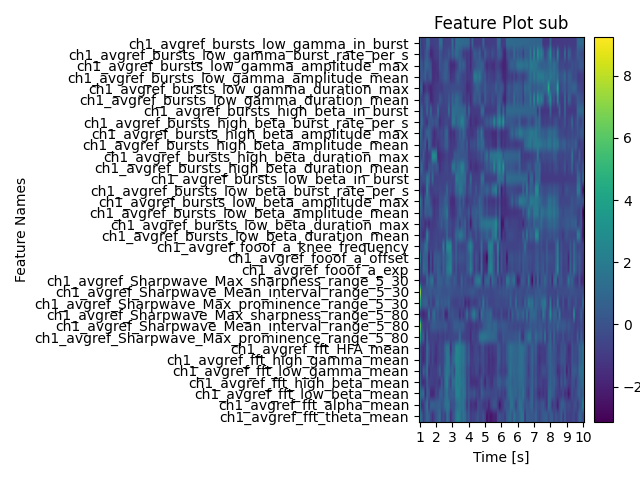
Seems like a lot of features were calculated. The time column tells us about each row time index. For the 6 specified channels, it is each 31 features. We can now use some in-built plotting functions for visualization.
Note
Due to the nature of simulated data, some of the features have constant values, which are not displayed through the image normalization.
analyzer.plot_all_features(ch_used="ch1")
nm.analysis.plot_corr_matrix(
figsize=(25, 25),
show_plot=True,
feature=analyzer.feature_arr,
)
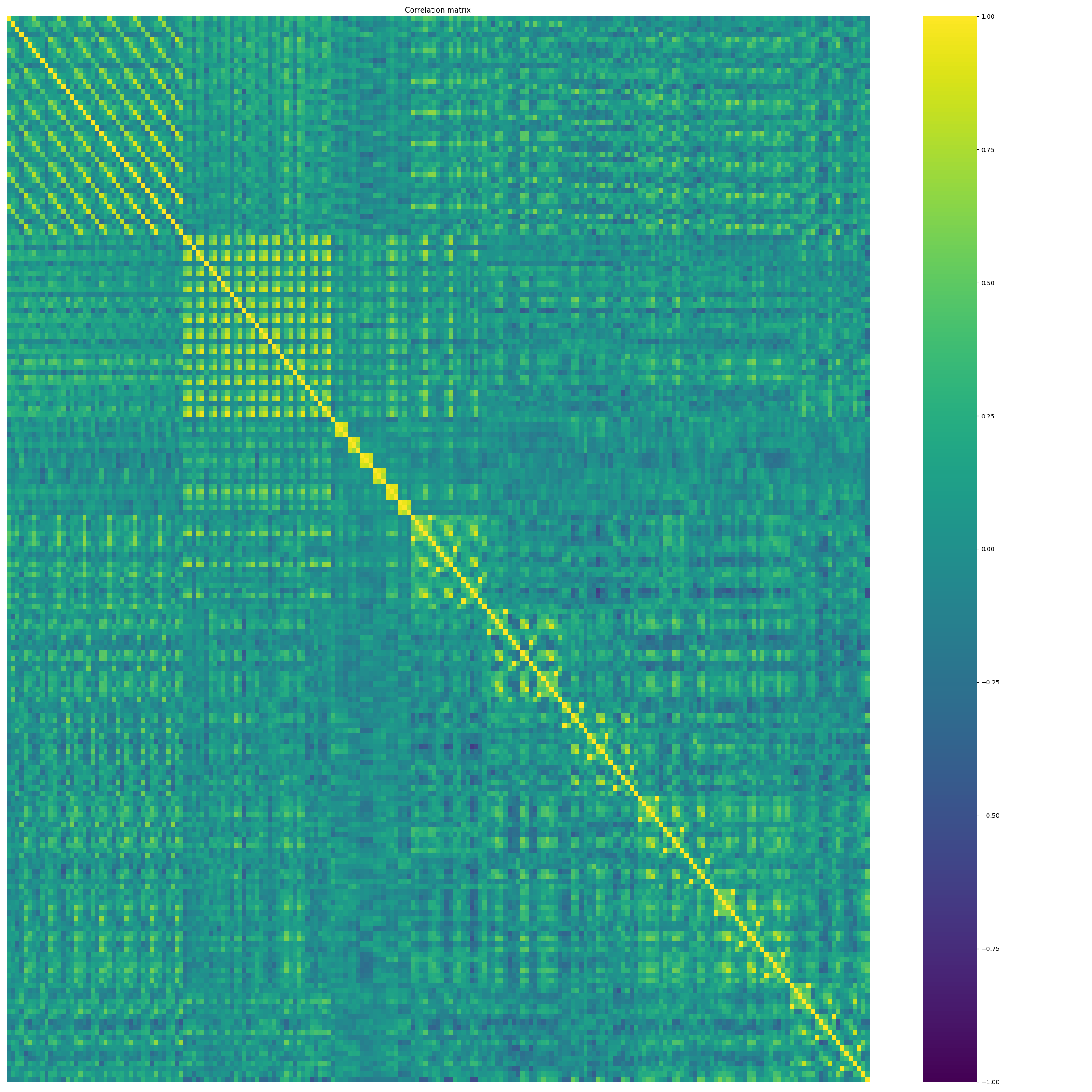
<Axes: title={'center': 'Correlation matrix'}>
The upper correlation matrix shows the correlation of every feature of every channel to every other. This notebook demonstrated a first demo how features can quickly be generated. For further feature modalities and decoding applications check out the next notebooks.
Total running time of the script: (0 minutes 14.498 seconds)